Head of the group:
Prof. Dr. Gábor Szabó

Staff:
Dr. László Imre

Dr. Péter Pál Nánási

Benhamza Ibtissem

Adél Vezendiné Nagy

Former members of the group:
- Katalin Goda, Ph.D.
- Tamás Varga, Ph.D.
- Zsolt Bacsó, M.D., Ph.D.
- György Fenyőfalvi, M.D.
- Lóránt Székvölgyi, Ph.D.
- Éva Hegedüs, Ph.D.
- Henrietta Nagy, Ph.D.
- Ferenc Fenyvesi, Ph.D.
- Pataki Judit, M.Sc.
- Szilágyi Ildikó, M.Sc.
- Erfaneh Firouzi Niaki, Dr. Pharm., Ph.D.
- Rosevalentine Bosire, Ph.D.
Former name of the group:
"Cell Biology of Anticancer Drug Action", which comprised also a section conducting membrane biophysics-oriented research on ABC transporters, that has become an independent group led by Katalin Goda.
Research:
We are investigating the complex relationships between nucleosome stability and organization of superhelical chromatin loops, also with the ambition of developing procedures for the epigenetic modulation of chromatin for therapeutic purposes.
Background:
Nucleosomal structure is repressive for transcription, hence eukaryotic gene regulation is primarily based on de-repression at the level of the chromatin. It is fine-tuned, among other factors, by histone posttranslational modifications (PTMs), and entails, and depends on, negative supercoiling of the DNA around the nucleosomal core. In line with this global picture, supercoil relaxation elicits a strong, PTM-dependent nucleosome destabilizing effect according to our published and unpublished data. Our previous work has also led us to recognize that DNA superhelicity has a strong and direct impact on the target size and binding affinity of two major classes of anticancer drugs: DNA intercalators and interstrand-crosslinking agents.
Current projects:
In a 3-decade long pursuit of the peculiar vulnerability of chromatin at loop-size intervals we have come to the working hypothesis that these phenomena are related to the presence of promoter-proximal endogeneous DNA breaks that may have a role in transcriptional regulation. We are investigating the mechanism of their generation, their possible functional role and their effect on higher-order chromatin structure, in S. cerevisiae and in human cells, including peripheral blood mononuclear cells.
Using our laser scanning cytometric assay of nucleosome stability (see below), we are investigating the molecular determinants of stability in the case of nucleosomes containing different PTMs or histone variants, with special interest in H2A.Z (see our manuscript deposited at https://www.biorxiv.org/content/10.1101/2021.02.22.432230v1).
Experimental systems:
Our experimental systems include mammalian cells, mouse embryonic stem cells and S. cerevisiae, studied by (a) confocal microscopic analysis of the localisation patterns of free 3’OHs detected by in situ nick translation or terminal transferase reaction relative to those of transcription related R-loops (RNA/DNA-hybrids), topoisomerase enzymes and epigenetic marks (PTMs); (b) a laser scanning cytometric (LSC) assay of nucleosome stability, measured in a PTM and cell cycle dependent manner; (c) an LSC assay of superhelical density and size distribution of loops; (d) molecular combing of DNA; (e,f) various gelelectrophoretic techniques (denaturing urea-agarose, CHEF), including single-cell electrophoresis of large DNA molecules; (g) a reverse South-Western blotting technique to map strand breaks and R-loops and (h) standard genomic techniques (microarray,chip seq).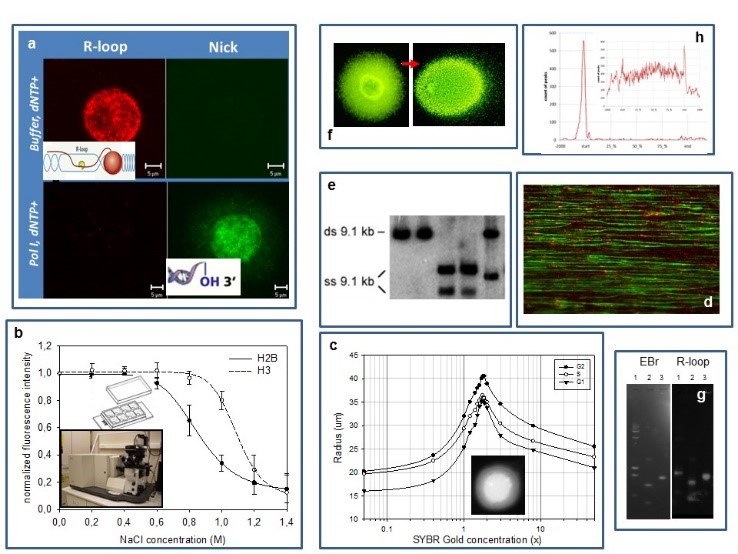
Representative publications:
- Firouzi Niaki E, Van Acker T, Imre L, Nánási P Jr, Tarapcsák S, Bacsó Z, Vanhaecke F, Szabó G. Interactions of Cisplatin and Daunorubicin at the Chromatin Level. Sci Rep. 2020 Jan 24;10(1):1107. doi: 10.1038/s41598-020-57702-7.
- Bosire R, Nánási P Jr, Imre L, Dienes B, Szöőr Á, Mázló A, Kovács A, Seidel R, Vámosi G, Szabó G.: Intercalation of small molecules into DNA in chromatin is primarily controlled by superhelical constraint. PLoS One. 2019 Nov 20;14(11):e0224936. doi: 10.1371/journal.pone.0224936. eCollection 2019.
- Nánási P Jr, Imre L, Firouzi Niaki E, Bosire R, Mocsár G, Türk-Mázló A, Ausio J, Szabó G.: Doxorubicin induces large-scale and differential H2A and H2B redistribution in live cells. PLoS One. 2020 Apr 16;15(4):e0231223. doi: 10.1371/journal.pone.0231223. eCollection 2020.
- Éva Hegedüs, Endre Kókai, Péter Nánási, László Imre, László Halász, Rozenn Jossé, Zsuzsa Antunovics, Martin R. Webb, Aziz El Hage, Yves Pommier, Lóránt Székvölgyi, Viktor Dombrádi and Gábor Szabó: Endogenous single-strand DNA breaks at RNA polymerase II promoters in Saccharomyces cerevisiae. Nucleic Acids Research, 2018 Nov 16;46(20):10649-10668. A summary of this publication is enclosed below.
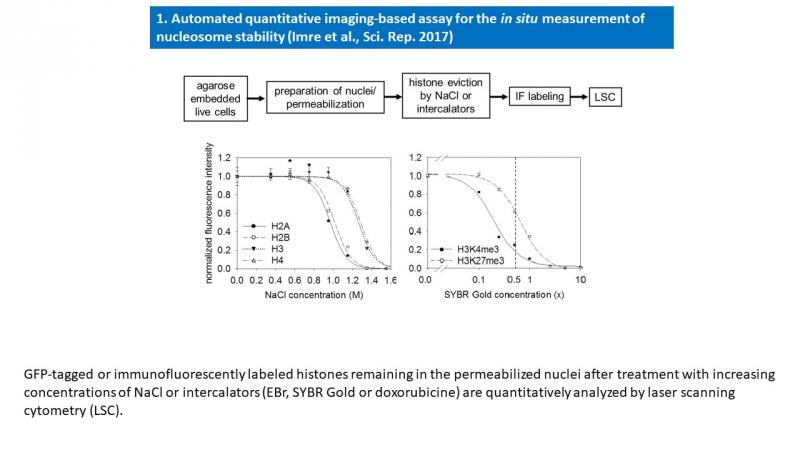
- Imre L, Simándi Z, Horváth A, Fenyőfalvi G, Nánási P, Niaki EF, Hegedüs É, Bacsó Z, Weyemi U, Mauser R, Ausio J, Jeltsch A, Bonner W, Nagy L, Kimura H, Szabó G. Nucleosome stability measured in situ by automated quantitative imaging. Sci Rep. 2017 Oct 6;7(1):12734.
- Imre L, Balogh I, Kappelmayer J, Szabó M, Melegh B, Wanker E, Szabó G. Detection of mutations by flow cytometric melting point analysis of PCR products. Cytometry A. 2011 Sep;79(9):720-6.
- Hegedüs E, Kókai E, Kotlyar A, Dombrádi V, Szabó G. Separation of 1-23-kb complementary DNA strands by urea-agarose gel electrophoresis. Nucleic Acids Res. 2009 Sep;37(17):e112.
- Hegedüs E, Imre L, Pataki J, Lizanecz E, Székvölgyi L, Fazakas F, Bacsó Z, Tóth A, Szabó M, Seres Z, Szabó G. Heteroduplex analysis using flow cytometric microbead assays to detect deletions, insertions, and single-strand lesions. Cytometry A. 2008 Mar;73(3):238-45.
- Székvölgyi L, Rákosy Z, Bálint BL, Kókai E, Imre L, Vereb G, Bacsó Z, Goda K, Varga S, Balázs M, Dombrádi V, Nagy L, Szabó G. Ribonucleoprotein-masked nicks at 50-kbp intervals in the eukaryotic genomic DNA. Proc Natl Acad Sci U S A. 2007 Sep 18;104(38):14964-9.
- Székvölgyi L, Bálint BL, Imre L, Goda K, Szabó M, Nagy L, Szabó G. Chip-on-beads: flow-cytometric evaluation of chromatin immunoprecipitation. Cytometry A. 2006 Oct 1;69(10):1086-91.
Summary of recent publications:
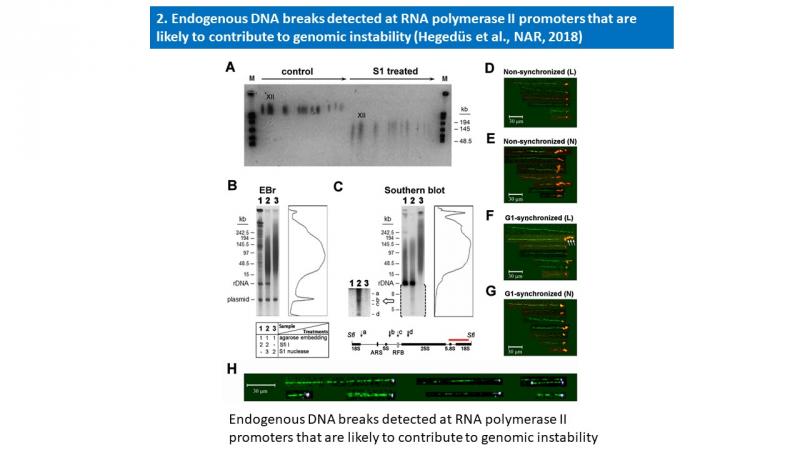
Molecular combing and gel electrophoretic studies revealed endogenous single-strand breaks, nicks with free 3′OH ends at ∼100 kb intervals in the genomic DNA of unperturbed and G1-synchronized Saccharomyces cerevisiae cells. These breaks accumulated at active RNA polymerase II promoters, reminiscent of the promoter-proximal transient DNA breaks of higher eukaryotes. Similar periodicity of endogenous nicks was found within the ribosomal rDNA cluster, involving every ∼10th of the tandemly repeated 9.1 kb units of identical sequence. Nicks were mapped by Southern blotting to a few narrow regions within the affected units. Three of them were overlapping the RNAP II promoters, while the region directing rDNA replication was spared of nicks. By using a highly sensitive method to map free DNA ends with 3′OH, they were shown to be distinct from other known rDNA breaks and linked to transcriptional regulation at the locus. Nicks were typically found at the ends of combed DNA molecules, occasionally together with R-loops, comprising a major pool of vulnerable sites that are connected with transcriptional regulation and are likely to contribute to genomic instability.
Support:
|
 |
 |
 |
|
|
|
|
OTKA(#1494); MAKA(91a.127); ETT (T-01 449/93); ETT (T-05 477/93); OTKA (T 17592); FEFA 1113; MKM 1062/1997; AKP 98-83 3,3; OTKA 032563; OMFB-02692/2000, 2001-2002; ALK-00036/2001, SBIR (NIH, USA), 2001-2002, ETT (614/2003), OTKA T 048742, NKFP 1A/041/04, GVOP - 3.1.1 - 2004 – 05 - 0440 /3.0, ETT 067/2006, OTKA 72762, TÁMOP-4.2.2-08/1/2008-0015, OTKA 101337; Marie Curie No. 292259, TAMOP 4.2.1/B-09/1/KONV-2010-0007, TÁMOP 4.2.2.A-11/1/KONV-2012-0023, Visegrad Fund 09/2012 StG, Debrecen University Symposium award 2013, GINOP-2.3.2-15-2016-00044, GINOP-2.3.3-15-2016-00020, OTKA K128770, K138524, COST CA15214, CA18127, Stipendium Hungaricum, Richter Gedeon Talentum Fund, Fulbright fellowship


