ABC proteins form one of the largest protein families and are present in every organism from bacteria to human. The majority of the 48 human ABC proteins are active pumps utilizing the energy of ATP hydrolysis for exporting various compounds out of cells. Interestingly, certain ABC transporters including P-glycoprotein (Pgp, ABCB1), and breast cancer resistance protein (BCRP, ABCG2) have extremely broad substrate spectra involving xenobiotics and numerous chemotherapeutic compounds applied in the treatment of various diseases (eg. antibiotics, reverse transcriptase inhibitors, antidepressants, narcotics and antineoplastic drugs). Thanks to their expression in tissue barriers including the intestinal epithelium, blood-brain, blood-placenta and blood-testis barriers as well as in drug metabolizing and drug extruding organs, e.g. in the liver and the kidney, Pgp and ABCG2 are important determinants of the pharmacokinetics of the above mentioned chemotherapeutic compounds. In addition, Pgp and ABCG2 are often expressed in tumor cells as well as in tumor stem cells. Since Pgp and ABCG2 can keep the concentration of cytotoxic drugs at sublethal level in the tumor cells, they are key players of chemotherapy resistance.
In view of the great physiological and medical importance of ABC transporters they are targets for pharmacological intervention. Detailed understanding of the working mechanism of ABC proteins is extremely important and might promote rational drug design. Our major aim is to understand how the ATP hydrolysis cycle is coupled to the different steps of substrate translocation process in various ABC proteins.
Methods:
In our laboratory we have a long tradition of studying the function of ABC transporters. In the past years we have studied the interaction of Pgp and ABCG2 with their substrates and inhibitors and examined the influence of the membrane microenvironment on the catalytic activity of Pgp. We have a long term experience with an anti-Pgp monoclonal antibody (UIC2), which reacts with a conformation sensitive extracellular epitope of the protein. In our previous work we applied this mAb for the in vitro and in vivo inhibition of Pgp and also exploited it for studying the conformational changes of Pgp in intact cells. Recently, using Staphylococcus aureus alpha-toxin-permeabilized cells we studied the conformational changes and substrate affinity changes of wild-type and catalytic site mutant Pgp molecules applying fluorescence based techniques.
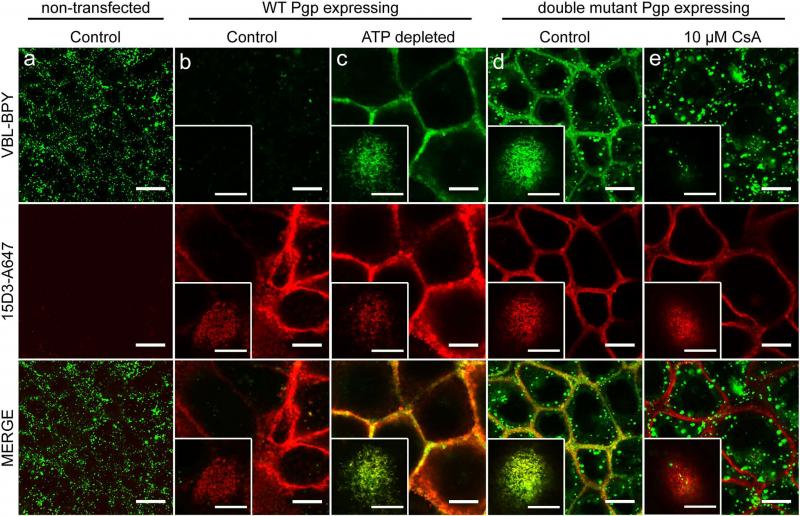
We have developed a simple confocal microscopy based method for describing the substrate affinity of Pgp at distinct steps of its catalytic cycle. Consistently with the Pgp-mediated efflux of vinblastine-bodipy (VBL-BPY), a Pgp substrate, cells expressing wild-type Pgp show significantly lower levels of intracellular fluorescence as compared to Pgp-negative cells when incubated with this fluorescent drug (panel b vs. a). ATP-depleted cells overexpressing wild-type Pgp sequester VBL-BPY in the plasma membrane (panel c). Pgp molecules are visualized by a fluorescently labeled anti-Pgp antibody (15D3-A647). Strikingly, plasma membrane sequestration of VBL-BPY was also observed in cells transfected with either single or double Walker A mutant Pgp variants (for double Walker A mutant see panel d). Addition of cyclosporine A (CsA), a competitive inhibitor of Pgp, prevented plasma membrane accumulation of VBL-BPY (panel e), suggesting that the enrichment of VBL-BPY in the plasma membrane is in each case due to its high-affinity binding to Pgp. To quantify the ratio of the substrate bound Pgps in the membrane, we calculate the Pearson’s cross-correlation coefficients between the channel of the VBL-BPY and the channel of the anti-Pgp antibody. Since the antibody staining was unchanged in the course of the treatments, the cross-correlation coefficients are determined by VBL-BPY binding to Pgp. If the cross-correlation coefficient is close to 1, all the Pgp molecules are in substrate binding conformation, if it is low (close to 0), Pgps do not bind VBL-BPY. (Bársony at al. Sci. Rep 6 (24810), 1-16., 2016)