In a 3-decade long pursuit of the peculiar vulnerability of chromatin at loop-size intervals we have come to the working hypothesis that these phenomena are related to the presence of promoter-proximal endogeneous DNA breaks that may have a role in transcriptional regulation. We are investigating the mechanism of their generation, their possible functional role and their effect on higher-order chromatin structure, in S. cerevisiae and in human cells, including peripheral blood mononuclear cells.
Using our laser scanning cytometric assay of nucleosome stability (see below), we are investigating the molecular determinants of stability int he case of nucleosomes containing different PTMs or histone variants, with special interest in H2A.Z.
We strive to make use of the cell biological perspective obtained on chromatin structure in anticancer drug research investigating the determinants of accessibility of small molecules to their target within the chromatin, by studying the interaction of DNA intercalators with chromatin in intact cells, by assessing the potentials of modulating superhelicity by intercalators on the toxicity of DNA cross-linking agents and their effect on nuclear architecture as reflected by the large-scale nuclear, nucleolar and nucleocytoplasmic reorganizations and relocations detectable by confocal microscopy.
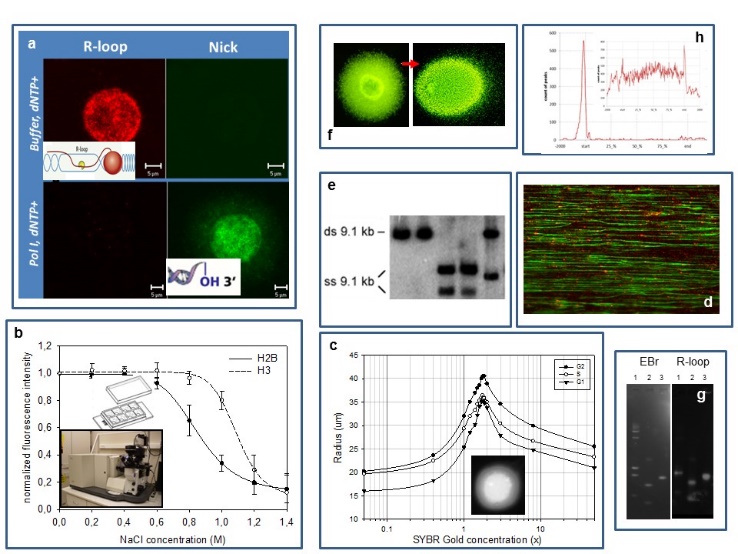
Our experimental systems include mammalian cells, mouse embryonic stem cells and S. cerevisiae, studied by (a) confocal microscopic analysis of the localisation patterns of free 3’OHs detected by in situ nick translation or terminal transferase reaction relative to those of transcription related R-loops (RNA/DNA-hybrids), topoisomerase enzymes and epigenetic marks (PTMs); (b) a laser scanning cytometric (LSC) assay of nucleosome stability, measured in a PTM and cell cycle dependent manner; (c) an LSC assay of superhelical density and size distribution of loops; (d) molecular combing of DNA; (e,f) various gelelectrophoretic techniques (denaturing urea-agarose, CHEF), including single-cell electrophoresis of large DNA molecules; (g) a reverse South-Western blotting technique to map strand breaks and R-loops and (h) standard genomic techniques (microarray,chip seq).